Resources
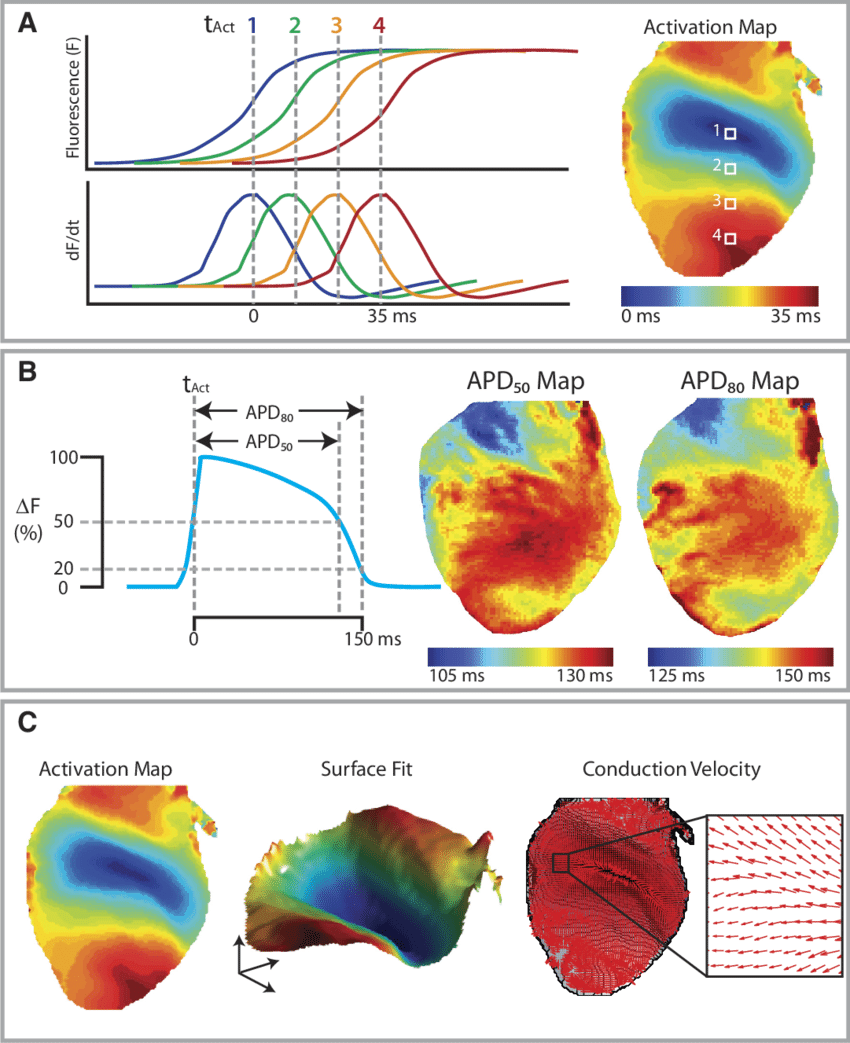
Optical Mapping
Background: Optical mapping is a technique we use to study cardiac electrophysiology.
Briefly, cardiac function is assessed by either using fluorescent dyes or intrinsic fluorescent
properties of the molecules in the heart. We routinely use voltage-sensitive fluorescent dyes
such as Di-4-ANEPPS, RH237 and Di-4-ANBDQBS that bind to the cell membrane of
cardiomyocytes. The fluorescence intensity of these dyes changes with the membrane potential.
High speed image acquisition of this optical signal from the heart gives an optical surrogate of
the cardiac action potential. Similarly, calcium indicator dyes such as Rhod2-AM is used to
record calcium transients from the heart. Additionally, NADH autofluorescence can also be
recorded using this technique as a measure of cardiac metabolic state.
Applications in the Efimov Lab: The Efimov Lab uses optical mapping techniques in many
forms ranging from single parametric-single field of view imaging to multiparametric-multiple
fields of view imaging and even panoramic approaches. Our lab is equipped with five optical
mapping systems, each specializing in specific applications:
1. High spatial resolution (256X256 pixels) single camera imaging.
2. Dual parameter optical mapping system for voltage and calcium recordings.
3. 3D printed dual optical mapping system for voltage/calcium/NADH imaging in upright
and horizontal camera orientation.
4. Panoramic optical mapping system.
5. Four camera optical mapping system for imaging multiple cardiac surfaces
simultaneously.
6. Triple imaging
A list of our recent publications using these systems can be found below.
Sharon A George, Jaclyn A Brennan, Igor R Efimov. Preclinical Cardiac Electrophysiology
Assessment by Dual Voltage and Calcium Optical Mapping of Human Organotypic Cardiac
Slices. JoVE, 2020. PMID: 32628156.Brianna Cathey, Sofian Obaid, Alexander M Zolotarev, Roman A Pryamonosov, Roman A
Syunyaev, Sharon A George, Igor R Efimov. Open-source multiparametric optical mapping. Sci
Rep, 2019. PMID: 30679527.Christopher Gloschat, Kedar Aras , Shubham Gupta, N. Rokhaya Faye, Hanyu Zhang , Roman A.
Syunyaev, Roman A. Pryamonosov, Jack Rogers, Matthew W. Kay, Igor R. Efimov. RHYTHM:
An Open Source Imaging Toolkit for Cardiac Panoramic Optical Mapping. Sci Rep, 2018. PMID:
29440763.Kedar K. Aras, Ndeye Rokhaya Faye, Brianna Cathey, Igor R. Efimov. Critical Volume of Human
Myocardium Necessary to Maintain Ventricular Fibrillation. Circ Arrhythm Electrophysiol,
2018. PMID: 30376733.Sharon A George, Igor R Efimov. Optocardiography: A review of its past, present, and future.
Curr Opin Biomed Eng, 2019. PMID: 31803858.Open-Source Resources: The Efimov Lab also provides open source resources for optical
mapping including analysis software and custom 3D printed optical mapping system
components design. These resources are available at https://github.com/optocardiography.
Rhythm 1.0: Rhythm 1.0 allows the user to display, condition and analyze optical mapping
signals recorded as .gsd or .rsd files using MiCam Ultima and MiCam 05 cameras from
SciMedia. This Matlab based GUI performs basic signal conditioning including filtering,
thresholding, binning, background removal, drift correction and normalization of the recorded
optical signals of the transmembrane potential. It also analyses the data to measure activation
times, conduction velocity, action potential duration, phase and dominant frequency.
Rhythm 1.2: Rhythm 1.2 is a newer version of this Matlab GUI that incorporates optical calcium
transient data analysis in addition to transmembrane potential analysis. Rhythm 1.2 performs
all the above mentioned signal conditioning and data analyses in addition to having the option
to visualize and analyze upto four data files simultaneously. This also allows to identify
simultaneously recorded signals and “link” the files for further analysis. For example, in
multiparametric analysis, simultaneously recorded voltage and calcium signals are “linked” to
indicate that these recordings were taken at the same time and in the same field of view.
Additionally, Rhythm 1.2 also has the capability to perform action potential and calcium
transient upstroke rise time analysis, calcium transient duration analysis and calcium decay
constant determination.
Rhythm 2.0: Rhythm 2.0 on the other hand is designed to analyze panoramic optical mapping
data of transmembrane potential. Users can generate 3D visualizations of different sized hearts
ranging from mouse to rabbit and project optical data onto the meshes using user-friendly
graphical user interfaces. This is useful in viewing how signals travel throughout the heart,
especially in arrhythmia studies.
3D printed multiparametric system: We recently developed a 3D printed optical mapping
system for multiparametric optical mapping with the possibility to image in the horizontal and
upright camera orientations. With the exclusion of lens, filters, dichroic mirrors and cameras, all
parts of this optical mapping system were custom-designed and 3D printed by our lab. The
components of this system included optical, mechanical and perfusion parts. Optical
components include filter cubes with adjustable and fixed dichroic mirror holders, camera
connectors and lens holders. Mechanical components include lab jack, mechanical lifts, camera
support cages and titling platforms to change system orientation (upright to horizontal).
Perfusion components include baths for Langendorff heart preparations (horizontal orientation)
as well as human slice preparations (upright orientation). All these designs (.stl files) are
available at Github (link above).
Contact us at paloma.amaral@northwestern.edu if you have questions.
Tissue Bank
Our lab has an extensive tissue bank consisting of over 400 donor and failing hearts (~40% females, ~60% males). Tissues are stored as flash frozen and in RNAlater at -80°C. Regions collected include:
· left atrial free wall
· left atrial appendage
· right atrial free wall
· right atrial appendage
· left ventricular base
· right ventricular base
· left ventricular apex
· right ventricular apex
· left ventricular adipose tissue
· right ventricular adipose tissue
· septum
Human heart program is very active, 13 papers were published for the period of 2020-2022 alone:
2. Electrophysiology and Arrhythmogenesis in the Human Right Ventricular Outflow Tract
3. Acetylcholine Reduces I Kr and Prolongs Action Potentials in Human Ventricular Cardiomyocytes
4. Chromatin Accessibility of Human Mitral Valves and Functional Assessment of MVP Risk Loci
5. Evidence of Superior and Inferior Sinoatrial Nodes in the Mammalian Heart
6. Heart slice culture system reliably demonstrates clinical drug-related cardiotoxicity
7. Genetic algorithm-based personalized models of human cardiac action potential
10. Fully implantable and bioresorbable cardiac pacemakers without leads or batteries
11. ZO-1 Regulates Intercalated Disc Composition and Atrioventricular Node Conduction
12. Identification of atrial fibrillation associated genes and functional non-coding variants
13. Elastic titin properties and protein quality control in the aging heart
Multiparametric slice culture platform for the investigation of human cardiac physiology
Human cardiac slices have recently emerged as a promising model of human cardiac physiology and pharmaceutical testing because it recapitulates organ level multi-cellular interactions and tissue architecture that traditional cell culture and animal models are not able to fully capture. Our lab has developed a method of collecting and culturing cardiac slices from human hearts rejected for organ transplant. To preserve the electrical and mechanical properties of these cardiac slices in long-term culture, we are developing an automated, microfluidic, fully-enclosed organotypic culture system that provides electrical, mechanical, and optical stimulation and real-time ECG monitoring to achieve long-term culture of cardiac slices.
Heart failure (HF) is a major cause of morbidity and mortality worldwide. The global burden of HF continues to rise, with prevalence rates estimated at 1-2% and incidence approaching 5-10 per 1000 persons annually. The complex pathophysiology of HF impacts virtually all aspects of normal cardiac function – from structure and mechanics to metabolism and electrophysiology – leading to impaired mechanical contraction and sudden cardiac death. Implantable cardioverter defibrillator (ICD) therapy remains the primary HF management strategy, as pharmacotherapy is generally ineffective. Thus, there is an acute need to translate basic research into improved HF therapy. Animal model investigations are a critical component of HF research. However, the translation from cellular and animal models to the bedside is hampered by significant differences between species and among physiological scales. Our studies over the last 8 years show that hypotheses generated in animal models need to be validated in human in vitro models. Importantly, however, human heart investigations can establish translational platforms for safety and efficacy studies before embarking on costly and risky clinical trials. Our research focusses on the human HF investigations of electrophysiology remodeling, metabolic remodeling, and β-adrenergic remodeling and on promising new technologies for HF research.